Samples
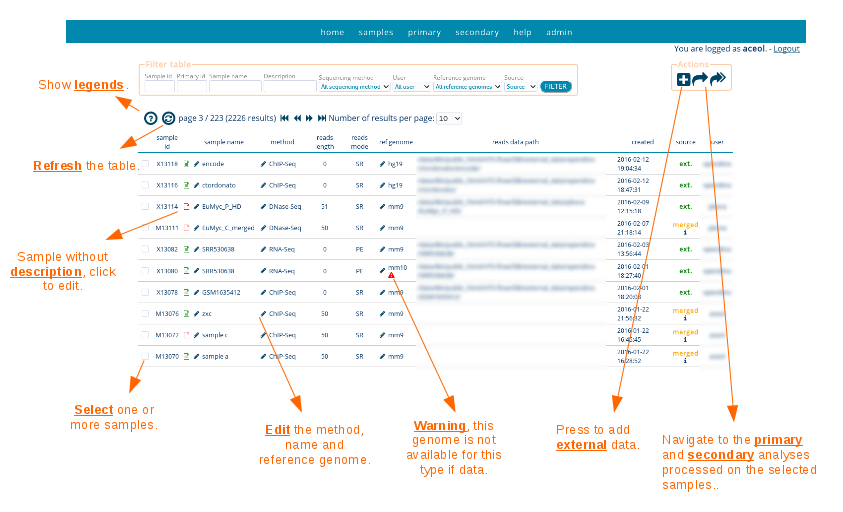
HTS-flow can be configured to import automatically samples from SMITH, a LIMS for handling next-generation sequencing workflows (http://cru.genomics.iit.it/smith/).
In addition, the samples can be imported (in Fastq format) directly by the user.
Finally, it is possible to import directly samples with their meta-data from GEO (http://www.ncbi.nlm.nih.gov/geo/). In this case, the user provides either a single expriment id (GSE), or a set of GSM ids. HTS-flow will take care at retrieving the metadata, downloading and preparing the raw data (generation of the fastq file), and creating new sample entries in the HTS-flow database.
There is no limitation in HTS-flow about the genomes that can be installed. The genome version need to be available at https://support.illumina.com/sequencing/sequencing_software/igenome.html. We provide a script that takes care about downloading the data, installating (or creating if it is not available) the R annotation file, and generating the genome data for teh different type of sequencing data.